Genetic analysis of major histocompatibility complex (MHC) Class I Exon 4-5 in cattle and buffalo using molecular and phylogenetic approaches
Keywords:
Bubalus bubalis, buffaloes, BoLA-A, BuLA-A typing, Exon 4-5, MHCAbstract
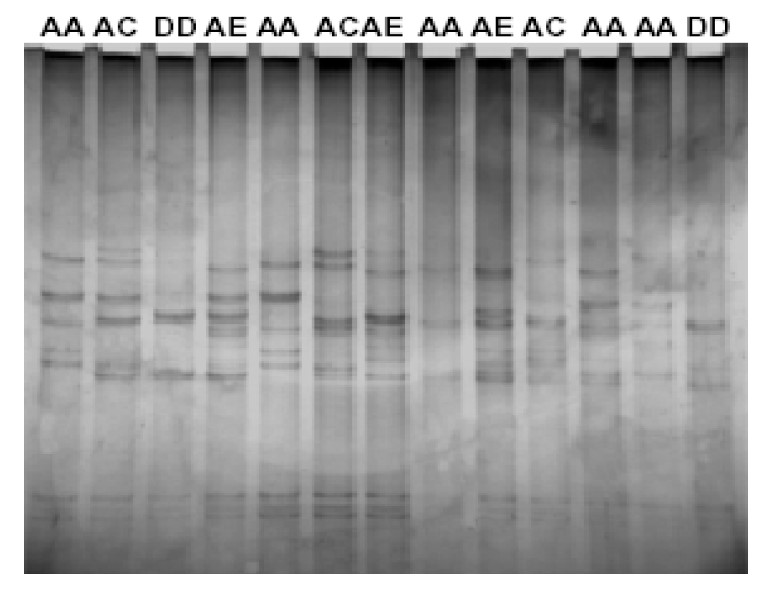
Characterization of major histocompatibility complex (MHC) Class I Exon 4-5 was carried out by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), single-strand confirmation polymorphism (SSCP) and sequencing techniques in crossbred cattle and buffaloes. Digestion of 559 bp fragment with HinfI and HaeIII restriction enzymes produced five and three distinct PCR-RFLP patterns in cattle and buffaloes, respectively. The genotype frequencies of HinfI patterns ranged from 0.015 to 0.456 in crossbred cattle, 0.080 to 0.600 in Mehsana and 0.050 to 0.500 in Bhadawari breeds of buffaloes. Similarly, the genotype frequencies of HaeIII patterns ranged from 0.045 to 0.636. The SSCP analysis showed that the BoLA-A and BuLA-A were highly polymorphic in cattle and buffaloes. A total of 10 alleles (AA, AB, BB, AC, AD, AE, AF, BC, DD and DB) were identified by PCR-SSCP. Four partial sequences, two each for crossbred cattle (AY790633 and AY790634) and buffaloes (AY785759 and AY785760), were submitted to the GenBank, NCBI. The sequencing results showed a number of amino acid changes in Exon 4-5 region. In conclusion, MHC Class I Exon 4-5 was found to be highly polymorphic in both cattle and buffaloes which may be exploited for association with traits of economic interest.
Downloads
Metrics
References
Behl, J.D., N.K. Verma, N. Tyagi, P. Mishra, R. Behl and B.K. Joshi. 2012. The major histocompatibility complex in bovines: A review. ISRN Veterinary Science, 2012: 1-12. DOI: 10.5402/2012/872710
Brunsberg, U., I. Edfors-Lilja, L. Andersson and K. Gustafsson. 1996. Structure and organization of pigMHC class IIDRB genes: Evidence for genetic exchange between loci. Immunogenetics, 44(1): 1-8. DOI: 10.1007/BF02602651
Castillo, S., V. Srithayakumar, V. Meunier and C.J. Kyle. 2010. Characterization of major histocompatibility complex (MHC) DRB Exon 2 and DRA Exon 3 fragments in a primary terrestrial rabies vector (Procyon lotor). PLoS One, 5(8): e12066. DOI: 10.1371/journal.pone.0012066
De, S. 2000. Molecular characterization of DRB exon2 in cattle and buffalo, Ph.D. Thesis, Deemed University, Indian Veterinary Research Institute, Izatnagar, India.
García, M.A.A., B.G. Yebra, A.L.L. Flores and E.G. Guerra. 2012. The major histocompatibility complex in transplantation. Journal of Transplantation, 2012: 842141. DOI: 10.1155/2012/842141
Li, D., K. Sun, Y. Zhao, A. Lin, S. Li, Y. Jiang and J. Feng. 2017. Polymorphism in the major histocompatibility complex (MHC Class II B) genes of the Rufous-backed Bunting (Emberiza jankowskii). PeerJ., 25(5): e2917. DOI: 10.7717/peerj.2917
Ng, J.H.J., M. Tachedjian, J. Deakin, J.W. Wynne, J. Cui, V. Haring, I. Broz, H. Chen, K. Belov, L.F. Wang and M.L. Baker. 2016. Evolution and comparative analysis of the bat MHC-I region. Sci. Rep.-UK, 15(6): 21256. DOI: 10.1038/srep21256
Sambrook, J. and D.W. Russell. 2001. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Pring Harbor Laboratory Press, New York, USA.
Sharma, A.K., B. Bhushan, S. Kumar, P. Kumar, A. Sharma and S. Kumar. 2004. Molecular characterization of Rathi and Tharparkar indigenous cattle (Bos indicus) breeds by RAPD-PCR. Asian Austral. J. Anim., 17(9): 1204-1209. DOI: 10.5713/ajas.2004.1204
Těšický, M. and M. Vinkler. 2015. Trans-species polymorphism in immune genes: General pattern or MHC-restricted phenomenon. J. Immunol. Res., 2015. DOI: 10.1155/2015/838035
Tizard, I. 2017. Veterinary Immunology-E-Book, 10th ed. Elsevier Health Sciences. The Major Histocompatibility Complex, Saunders, Country, p. 101-112.
Van Den Bussche, R.A., S.R. Hoofer and R.L. Lochmiller. 1999. Characterization of Mhc-DRB allelic diversity in white-tailed deer (Odocoileus virginianus) provides insight into Mhc-DRB allelic evolution within Cervidae. Immunogenetics, 49(5): 429-437. DOI: 10.1007/s002510050516
Wieczorek, M., E.T. Abualrous, J. Sticht, M. Álvaro-Benito, S. Stolzenberg, F. Noé and C. Freund. 2017. Major histocompatibility complex (MHC) Class I and MHC Class II proteins: Conformational plasticity in antigen presentation. Front. Immunol., 8(292). DOI: 10.3389/fimmu.2017.00292




.png)








