Molecular detection of multidrug resistant Escherichia coli isolated from buffalo meat and offal
Keywords:
Bubalus bubalis, buffalo meat, offal, E. coli, multidrug resistance, virulence genesAbstract
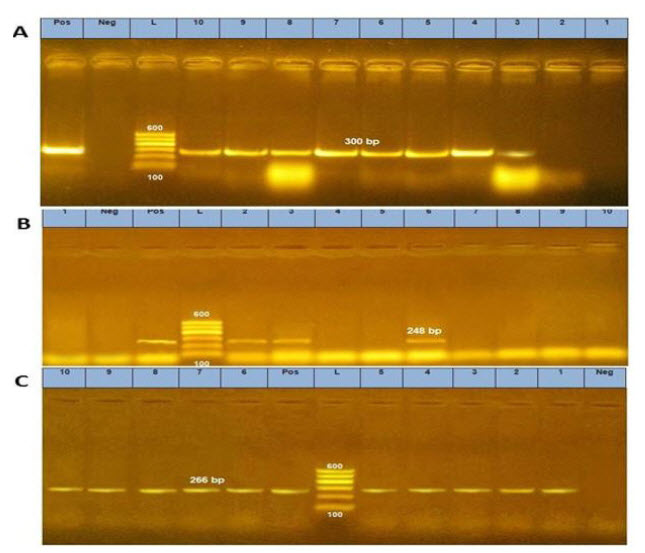
Water buffalo (Bubalus bubalis) is a significant source of high-quality milk and meat in Egypt. The goal of this research was to investigate the serotypes, virulence genes, and antibiotic resistance profiles of Escherichia coli (E. coli) detected in buffalo meat and offal. E. coli was detected at 26.67%, 56.67%, 60%, 13.33%, and 23.33% of buffalo meat, minced meat, liver, heart, and kidney, respectively. The detected E. coli was serologically identified as enteropathogenic E. coli (EPEC) O78, O2:H6, O1:H7, O153:H2 and O153:H11. Enterohemorrhagic E. coli (EHEC) O91: H21 and O26: H11 in addition to enterotoxigenic E. coli (ETEC) O127:H6, O126:H11, and O128:H2. All of the isolated E. coli was resistant to penicillin. Furthermore, 85%, 77.5%, 72.5%, 60%, 55%, and 52.5% of the isolates were resistant to erythromycin, ampicillin, cephalothin, nalidixic acid, kanamycin, and oxacillin, respectively. Thirty-one out of forty examined E. coli (77.5%) were resistant to at least three antibiotic classes and thus were considered as multidrug resistant (MDR) E. coli. Ten MDR isolates were submitted for molecular determination of virulence genes including iron uptake (iutA) gene, attaching and effacing (eae) gene, and increased serum survival (iss) gene, with detection rates of 80%, 30%, and 100%, respectively. In conclusion, buffalo meat and offal can be considered as potential sources of multidrug resistant E. coli.
Downloads
Metrics
References
Anas, M. and A. Malik. 2021. Impact of sodium alginate packaging film synthesized using Syzygium cumini seed extract on multi drug resistant Escherichia coli isolated from Raw buffalo meat. Indian J. Microbiol., 61(2): 137-150. DOI: 10.1007/s12088-021-00923-9
American Public Health Association, (APHA). 2001. Compendium of Methods for the Microbiological Examination of Foods, 4th ed. American Public Health Association, Washington DC., USA.
Aslam, M. and C. Service. 2006. Antimicrobial resistance and genetic profiling of Escherichia coli from a commercial beef packing plant. J. Food Protect., 69(7): 1508-1513. DOI: 10.4315/0362-028x-69.7.1508
Bantawa, K., S.N. Sah, D.S. Limbu, P. Subba and A. Ghimire. 2019. Antibiotic resistance patterns of Staphylococcus aureus, Escherichia coli, Salmonella, Shigella and Vibrio isolated from chicken, pork, buffalo and goat meat in eastern Nepal. BMC Research Notes, 12(1): 1-6. DOI: 10.1186/s13104-019-4798-7
Barlow, R.S., K.S. Gobius and P.M. Desmarchelier. 2006. Shiga toxin-producing Escherichia coli in ground beef and lamb cuts: Results of a one-year study. Int. J. Food Microbiol., 111(1): 1-5. DOI: 10.1016/j.ijfoodmicro.2006.04.039
Barlow, R.S., R.G. Hirst, R.E. Norton, C. Ashhurst-Smith and K.A. Bettelheim. 1999. A novel serotype of enteropathogenic Escherichia coli (EPEC) as a major pathogen in an outbreak of infantile diarrhoea. J. Med. Microbiol., 48(12): 1123-1125. DOI: 10.1099/00222615-48-12-1123
Bélanger, L., A. Garenaux, J. Harel, M. Boulianne, E. Nadeau and C.M. Dozois. 2011. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Mic., 62(1): 1-10. DOI: 10.1111/j.1574-695X.2011.00797.x
Biswas, A.K., N. Kondaiah, K.N. Bheilegaonkar, A.S.R. Anjaneyulu, S.K. Mendiratta, C. Jana and R.R. Kumar. 2008. Microbial profiles of frozen trimmings and silver sides prepared at Indian buffalo meat packing plants. Meat Sci., 80(2): 418-422. DOI: 10.1016/j.meatsci.2008.01.004
Christopher, A.F., S. Hora and Z. Ali. 2013. Investigation of plasmid profile, antibiotic susceptibility pattern multiple antibiotic resistance index calculation of Escherichia coli isolates obtained from different human clinical specimens at tertiary care hospital in Bareilly-India. Annals of Tropical Medicine and Public Health, 6(3): 285-291. DOI: 10.4103/1755-6783.120985
Clinical Laboratory Standards Institute, CLSI. 2015. Performance Standards for Antimicrobial Susceptibility Testing: Twentyfifth Informational Supplement. Wayne: Clinical Laboratory Standards Institute. Available on: https://www.facm.ucl.ac.be/intra net/CLSI/CLSI-2015- M100-S25-original.pdf.
Cyoia, P.S., G.R. Rodrigues, E.K. Nishio, L.P. Medeiros, V.L. Koga, A.P.D. Pereira and R.K. Kobayashi. 2015. Presence of virulence genes and pathogenicity islands in extraintestinal pathogenic Escherichia coli isolates from Brazil. J. Infect. Dev. Countr., 9(10): 1068-1075. DOI: 10.3855/jidc.6683
Elabbasy, M.T., M.A. Hussein, F.D. Algahtani, A. El-Rahman, I. Ghada, A.E. Morshdy and A.A. Adeboye. 2021. MALDI-TOF MS based typing for rapid screening of multiple antibiotic resistance E. coli and virulent non-O157 shiga toxin-producing E. coli isolated from the slaughterhouse settings and beef carcasses. Foods, 10(4): 820. DOI: 10.3390/foods10040820
FAO, Food and Agriculture Organization. 1991. World and Health Organization Surveillance Program for Central of Food Borne Infection and Intoxication in Europe. Food and Agriculture Organization CCRTFHZFIHCerinaryicine (BGVV), Berlin, Germany.
Garrity, G.M., D.J. Brennner, N.R. Krieg and J.T. Staley. 2005. Bergey’s manual of systematic bacteriology. The Proteobacteria, the Gammaproteobacteria, 2nd ed. Springer, Berlin, Germany.
Hanan, E., A. Amal and I. Helal. 2020. Molecular detection of some virulence genes of e. coli isolates from meat and meat product in port-said governorate. Assiut Veterinary Medical Journal, 66(167): 1-11. DOI: 10.21608/avmj.2020.167029
Hill, W.E., R. Suhalim, H.C. Richter, C.R. Smith, A.W. Buschow and M. Samadpour. 2011. Polymerase chain reaction screening for Salmonella and enterohemorrhagic Escherichia coli on beef products in processing establishments. Foodborne Pathog. Dis., 8(9): 1045-1053. DOI: 10.1089/fpd.2010.0825
Jørgensen, S.L., M. Stegger, E. Kudirkiene, B. Lilje, L.L. Poulsen, T. Ronco, T.P. Dos Santos, K. Kiil, M. Bisgaard, K. Pedersen, L.K. Nolan, L.B. Price, R.H. Olsen, P.S. Andersen and H. Christensen. 2019. Diversity and population overlap between avian and human Escherichia coli belonging to sequence type 95. MSphere, 4(1). DOI: 10.1128/mSphere.00333-18
Kaper, J.B., J.P. Nataro and H.L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol., 2(2): 123-140. DOI: 10.1038/nrmicro818
Kok, T., D. Worswich and E. Gowans. 1996. Some serological techniques for microbial and viral infections. In Collee, J., A. Fraser, B. Marmion and A. Simmons (eds.) Practical Medical Microbiology Churchill Livingstone, Edinburgh, UK.
Kolář, M., K. Urbánek and T. Látal. 2001. Antibiotic selective pressure and development of bacterial resistance. Int. J. Antimicrob. Ag., 17(5): 357-363. DOI: 10.1016/s0924-8579(01)00317-x
López-Banda, D.A., E.M. Carrillo-Casas, M. Leyva-Leyva, G. Orozco-Hoyuela, Á.H. Manjarrez-Hernández, S. Arroyo-Escalante and R. Hernández-Castro. 2014. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed. Res. Int., 1: 1-10. DOI: 10.1155/2014/959206
Molina-López, J., G. Aparicio-Ozores, R.M. Ribas-Aparicio, S. Gavilanes-Parra, M.E. Chávez-Berrocal, R. Hernández-Castro and H.Á. Manjarrez-Hernández. 2011. Drug resistance, serotypes, and phylogenetic groups among uropathogenic Escherichia coli including O25-ST131 in Mexico City. J. Infect. Dev. Countr., 5(12): 840-849. DOI: 10.3855/jidc.1703
Morshdy, A.E., A.I. El-Atabany, M.A. Hussein and W.S. Darwish. 2013. Oxytetracycline residues in bovine carcasses slaughtered at Mansoura Abattoir, Egypt. Jpn. J. Vet. Res., 61(Suppl.): S44-S47.
Oliveira, E.S., M.V. Cardozo, M.M. Borzi, C.A. Borges, E.A.L. Guastalli and F.A. Ávila. 2019. Highly pathogenic and multidrug resistant avian pathogenic Escherichia Coli in free-range chickens from Brazil. Revista Brasileira de Ciencia Avicola, 21(1): 1-8. DOI: 10.1590/1806-9061-2018-0876
Österblad, M., A. Hakanen, R. Manninen, T. Leistevuo, R. Peltonen, O. Meurman and P. Kotilainen. 2000. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. Antimicrob. Agents Ch., 44(6): 1479-1484. DOI: 10.1128/aac.44.6.1479-1484.2000
Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: Characterization of a new intimin variant. Infect. Immun., 68(1): 64-71. DOI: 10.1128/IAI.68.1.64-71.2000
Rahman, M. A., Rahman, A. K. M. A., M.A. Islam and M.M. Alam. 2017. Antimicrobial resistance of Escherichia coli isolated from milk, beef and chicken meat in Bangladesh. Bangladesh Journal of Veterinary Medicine, 15(2): 141-146.
Schjørring, S. and K.A. Krogfelt. 2011. Assessment of bacterial antibiotic resistance transfer in the gut. International Journal of Microbiology, 1: 1-10. DOI: 10.1155/2011/312956
Shekh, C.S., V.V. Deshmukh, R.N. Waghamare, N.M. Markandeya and M.S. Vaidya. 2013. Isolation of pathogenic Escherichia coli from buffalo meat sold in Parbhani city, Maharashtra, India. Vet. World, 6(5): 277-279.
Van Boeckel, T.P., C. Brower, M. Gilbert, B.T. Grenfell, S.A. Levin, T.P. Robinson and R. Laxminarayan. 2015. Global trends in antimicrobial use in food animals. In Proceedings of the National Academy of Sciences of the United State of America, 112(18): 5649-5654. DOI: 10.1073/pnas.1503141112
Wandersman, C. and I. Stojiljkovic. 2000. Bacterial heme sources: The role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol., 3(2): 215-220. DOI: 10.1016/s1369-5274(00)00078-3
Wang, F., L. Jiang, Q. Yang, W. Prinyawiwatkul and B. Ge. 2012. Rapid and specific detection of Escherichia coli serogroups O26, O45, O103, O111, O121, O145, and O157 in ground beef, beef trim, and produce by loop-mediated isothermal amplification. J. Appl Environ Microbiol, 78(8), 2727-2736. DOI: 10.1128/AEM.07975-11
Wang, G., C.G. Clark and F.G. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol., 40(10): 3613-3619. DOI: 10.1128/JCM.40.10.3613-3619.2002
Yaguchi, K., T. Ogitani, R. Osawa, M. Kawano, N. Kokumai, T. Kaneshige, T. Noro, K. Masubuchi and Y. Shimizu. 2007. Virulence Factors of Avian Pathogenic Escherichia coli Strains Isolated from Chickens with Colisepticemia in Japan. Avian Dis., 51(3): 656-662. DOI: 10.1637/0005-2086(2007)51[656:VFOAPE]2.0.CO;2
Ziauddin, K.S., N.S. Mahendrakar, D.N. Rao, B.S. Ramesh and B.L. Amla. 1994. Observations on some chemical and physical characteristics of buffalo meat. Meat Sci., 37(1): 103-113. DOI: 10.1016/0309-1740(94)90148-1




.png)








